Translational Omics Program
Genomically powered medicine
An estimated 25 million Americans are living with rare diseases, the majority of which are genetic and undiagnosed. Genomically powered individualized medicine is transforming the ability to diagnose and treat these patients and prevent future disease.
State-of-the-art clinical testing is generating a wealth of genomics data. This enables patients to find answers to their diagnostic dilemmas and in some cases leads to new therapeutic opportunities. Sequencing large patient populations is showing that genomic results are widely applicable and highly impactful in clinical care decisions, allowing medical professionals to offer patients more predictive and preventative medicine. In these ways and more, genomics is transforming patient care and improving patients' lives.
Translational Omics Program
As genomics increasingly becomes a standard component of individualized medical care, the challenges around data interpretation grow. The Center for Individualized Medicine established the Translational Omics Program in 2015 to address this challenge by providing post-analytical interpretive services for omics-related clinical and translational research activities. The program studies rare and undiagnosed genetic disease, preventive genomic screening, and pre-myeloid disease testing.
The Translational Omics Program has a rich, multidisciplinary team environment focused on developing the workforce of the future by training postdoctoral fellows, graduate students and interns. The program also provides translational omics interpretative services through its institutional recharge facility. Team members develop and implement cutting-edge methods for interpretation of omics data to facilitate patient diagnoses and treatment. This is accomplished through computational data review, data integration and data interpretation services as well as traditional laboratory in vitro and in vivo functional studies of genetic findings.
Translation Omics Program team members provide services including:
- Characterization of variants of uncertain significance and variants in genes of uncertain significance
- Execution and interpretation of complementary omics profiling, such as RNA sequencing, whole-genome sequencing and methylation sequencing
- In silico protein modeling
- Functional laboratory studies
- Identification or creation of collaborations and patient cohort studies
- Manuscript development and support
If you are interested in a fellowship or want to learn more about our team, email the Translation Omics Program Recharge Facility at TOPRF@mayo.edu to learn more about how we can support you.
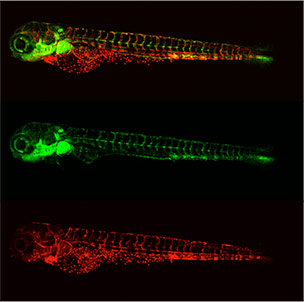
Larval transgenic zebrafish expressing green fluorescent protein in the vasculature and red fluorescent protein in the blood; Tg(fli:eGFP/gata-2:dsRed)
Projects
Exome sequencing for patients with rare and undiagnosed diseases has provided diagnostic answers for thousands of patients who previously lacked a clear explanation for their symptoms. This testing has become so powerful that institutions across the world, including Mayo Clinic, have adopted this approach to testing patients with suspected rare genetic diseases.
Despite the amazing impact this testing has had on some patients, only about 25% find a diagnosis with the current approach. This means roughly three-fourths of the patients tested fail to receive a genetic diagnosis.
The goal of this project is to re-evaluate clinically generated sequencing data, pushing beyond clinically reported results to identify new variants that may explain the disease. The research team then designs and carries out additional studies to evaluate these variants' abilities to cause diseases, or their pathogenicity.
The goal of the RNA-Assisted Diagnostics and Novel Therapeutics (RADiaNT) project is to transform Mayo Clinic's care for patients with rare and undiagnosed genetic diseases by implementing complementary RNA sequencing to improve the quantity and quality of diagnoses and point to individualized therapeutics.
As part of this project, more than 250 patients with rare and undiagnosed genetic diseases who have had inconclusive clinical exome or genome sequencing will receive RNA sequencing at Mayo Clinic. Researchers are using bioinformatics to critically analyze these data and strategically combine multiple analyses "pipelines" into a cohesive framework.
Additionally, in the initial phase of the RADiaNT project, researchers will select a small number of successfully diagnosed patients to receive personalized antisense oligo (ASO) therapeutics for evaluation in pilot studies. The goal of this research is to assess the feasibility of using ASOs to treat rare genetic diseases and expand the options for individualized treatments.
Clinicians are increasingly using DNA and RNA sequencing to diagnose patients with suspected but undiagnosed Mendelian diseases, which are rare genetic conditions.
However, many of these complex cases require a more comprehensive understanding of the molecular causes of a patient's disease, which incorporates epigenetic regulation. Hereditary genetic diseases can be caused by errors in DNA methylation, which cause changes in the expression of genes related to the disease.
To better understand and diagnose rare genetic diseases, Mayo Clinic researchers are performing methylation sequencing for 100 patients with rare and undiagnosed genetic diseases. Then, the researchers integrate the results with the patients' existing DNA and RNA sequencing to create multi-omic profiles.
Whole-exome sequencing (WES) has transformed genetic diagnosis. However, for rare and undiagnosed diseases, only 25% to 50% of patients receive confirmed diagnoses, depending in part on how much prior testing they've had. A majority of patients remain undiagnosed after exome sequencing or are told they have a variant of uncertain significance (VUS).
However, researchers across the field of exome sequencing report findings about new variations in scientific publications and collect it in databases every day. Consequently, any patient's diagnosis of VUS could — at any time — be reclassified by emerging findings, turning previously unresolved tests into diagnostic answers.
Due to the high number of unsolved cases, manual re-analysis is unrealistic; no clinical lab is known to be routinely performing this task. However, the Mayo Clinic Center for Individualized Medicine's Re-Analysis of Negative WES (RENEW) project is using an automated bioinformatics system that identifies new findings in three of the major genetic databases — ClinVar, the Human Gene Mutation Database and Online Mendelian Inheritance in Man.
Researchers then use this information to automatically re-annotate genomic data from undiagnosed patients and identify test results warranting further review. Using additional filters that account for the inheritance pattern and population frequency of a given disease, they can focus on relevant information and re-analyze inconclusive tests in minutes.
The Translational Omics Program is developing extensive annotation that uses recent scientific publications to interpret genetic variations as effectively and accurately as possible. This includes developing bioinformatics software and application programming interfaces for scalable and efficient variant interpretation of large-scale population screening initiatives.
At the crux of today's advanced clinical genetic testing is a massive data analysis need, as each individual patient has tens of thousands of unique genetic changes. To understand these data, scientists must integrate them with a detailed account of the patient's condition or phenotype and with existing scientific knowledge on human health genetics.
To address this need, the Translational Omics Program uses an intuitive computer system to capture clinicians' observations about patients and store them in a specific way to facilitate automated analysis. These specially stored data are called structured phenotype data.
Program researchers also are implementing a process to automatically cross-reference these structured phenotype data with the published scientific literature, existing biological and clinical databases, and certain basic-science data repositories to quickly identify the set of genes related to a patient's observed condition.
Finally, the team uses machine learning tools to integrate patient-specific genetic changes identified by whole-exome sequencing with the existing scientific literature. This system can identify genetic changes that are likely related to a patient's disease.
Current testing for rare and undiagnosed diseases evaluates a patient's DNA within those regions of their genome that contain genes. The majority of current clinical knowledge about the genetics of disease is focused on these regions.
Another type of genomic data, regarding a patient's RNA, is a measure of which genes are currently active in the patient. Studying both the changes to a patient's genome (DNA) and the activity of the patient's genes (RNA) helps researchers better understand the potential ramifications of specific genetic changes.
To this end, scientists in the Translational Omics Program are evaluating the use of RNA sequencing, along with several other types of genomic testing, to complement the findings obtained from the whole-exome sequencing. The team has discovered that in a subset of patients, this data integration can greatly improve diagnostic capabilities and help identify the underlying cause of a patient's genetic disease.
A significant challenge with current genetic testing is the large number of genetic variants of uncertain significance (VUSs) that are identified. These are genetic changes that are poorly studied, or are identified in genes for which little is known. Consequently, clinical interpretation of the genetic change is extremely difficult.
To better understand a subset of these VUSs, the Translational Omics Program has established a functional studies initiative using protein and animal models to complement laboratory testing. Protein modeling allows researchers to predict and visualize the impact a genetic variant has on a patient's protein, leading to proposed experimental tests to evaluate its subsequent biological impacts.
The team also is using cutting-edge genome engineering technologies to introduce a patient's genetic variant into an animal model or lab system. This allows researchers to make observations and carry out tests to better understand the functional impact of the genetic change.
Program leader
Faculty
- Joseph (Joe) D. Farris, Ph.D.
- Johannes (Jan) M. Verheijen, Ph.D.
- Matheus Vernet Machado Bressan Wilk, M.D.
- Nancy William, Ph.D.
- Patrick R. Blackburn, Ph.D. — Transitioned into Mayo Clinic's clinical Laboratory Genetics and Genomics Fellowship
- Nicole J. Boczek, Ph.D. — Transitioned into Mayo Clinic's clinical Laboratory Genetics and Genomics Fellowship
- Margot A. Cousin, Ph.D. — Transitioned into an academic research position
- Alejandro Ferrer, Ph.D. — Transitioned into an academic research position
- Aditi Gupta, Ph.D. — Transitioned into a second postdoc fellowship with a cancer focus
- Charu Kaiwar, M.D., Ph.D. — Transitioned into a genetic testing industry staff position
- Erica L. Macke, Ph.D. — Transitioned into Mayo Clinic's clinical Laboratory Genetics and Genomics Fellowship
- Joel A. Morales Rosado, M.D. — Transitioned into a pathology residency
- Rory J. Olson, Ph.D. — Transitioned into a clinical variant scientist position
- Filippo Pinto e Vairo, M.D., Ph.D. — Transitioned into an academic research position
- Stephanie L. Safgren, Ph.D. — Transitioned into an oncology research position
- Laura E. Schultz-Rogers, Ph.D. — Transitioned into Mayo Clinic's clinical Laboratory Genetics and Genomics Fellowship
- Jennifer Arroyo, Ph.D. — Clinical Variant Scientist
- Lindsay A. Mulvihill, CCRP — Senior Program Coordinator, Research
- Rory J. Olson, Ph.D. — Clinical Variant Scientist
- Carolyn (Caer) R. Vitek, Ed.D., M.S. — Operations Manager
- Karl J. Clark, Ph.D. — Associate Consultant II
- Maria C. O'Connell — Research Technologist
- Christopher (Chris) T. Schmitz — Senior Research Technologist
- Andrew C. (Collin) Osborne, M.S. — Bioinformatician
These collaborators work with the Translational Omics Program through a formal collaboration with Medical College of Wisconsin:
- Raul A. Urrutia, M.D. — Director of the Human and Molecular Genetics Center and Professor, Department of Surgery
- Michael T. Zimmermann, Ph.D. — Assistant Professor, Clinical and Translational Science Institute
Publications
Collectively, Mayo Clinic authors publish more than 5,000 articles a year in biomedical journals.
Publishing in medical journals is an expected scholarly activity of professional practice and aligns with our value of sharing expertise and best practices to facilitate the advancement of medical practice worldwide.
Featured research articles
Read peer-reviewed articles about key findings from the Translational Omics Program:
- Blackburn PR, Xu Z, Tumelty KE, Zhao RW, Monis WJ, Harris KG, Gass JM, Cousin MA, Boczek NJ, Mitkov MV, Cappel MA, Francomano CA, Parisi JE, Klee EW, Faqeih E, Alkuraya FS, Layne MD, McDonnell NB, Atwal PS. Bi-allelic Alterations in AEBP1 Lead to Defective Collagen Assembly and Connective Tissue Structure Resulting in a Variant of Ehlers-Danlos Syndrome. The American Journal of Human Genetics. 2018; doi:10.1016/j.ajhg.2018.02.018.
- Cousin MA, Creighton BA, Breau KA, Spillmann RC, Torti E, Dontu S, Tripathi S, Ajit D, Edwards RJ, Afriyie S, Bay JC, Harper KM, Beltran AA, Munoz LJ, Falcon Rodriguez L, Stankewich MC, Person RE, Si Y, Normand EA, Blevins A, May AS, Bier L, Aggarwal V, Mancini GMS, van Slegtenhorst MA, Cremer K, Becker J, Engels H, Aretz S, MacKenzie JJ, Brilstra E, van Gassen KLI, van Jaarsveld RH, Oegema R, Parsons GM, Mark P, Helbig I, McKeown SE, Stratton R, Cogne B, Isidor B, Cacheiro P, Smedley D, Firth HV, Bierhals T, Kloth K, Weiss D, Fairley C, Shieh JT, Kritzer A, Jayakar P, Kurtz-Nelson E, Bernier RA, Wang T, Eichler EE, van de Laar IMBH, McConkie-Rosell A, McDonald MT, Kemppainen J, Lanpher BC, Schultz-Rogers LE, Gunderson LB, Pichurin PN, Yoon G, Zech M, Jech R, Winkelmann J; Undiagnosed Diseases Network; Genomics England Research Consortium, Beltran AS, Zimmermann MT, Temple B, Moy SS, Klee EW, Tan QK, Lorenzo DN. Pathogenic SPTBN1 variants cause an autosomal dominant neurodevelopmental syndrome. Nature Genetics. 2021; doi:10.1038/s41588-021-00886-z.
- Klee EW, Cousin MA, Pinto E Vairo F, Morales-Rosado JA, Macke EL, Jenkinson WG, Ferrer A, Schultz-Rogers LE, Olson RJ, Oliver GR, Sigafoos AN, Schwab TL, Zimmermann MT, Urrutia RA, Kaiwar C, Gupta A, Blackburn PR, Boczek NJ, Prochnow CA, Lowy RJ, Mulvihill LA, McAllister TM, Aoudia SL, Kruisselbrink TM, Gunderson LB, Kemppainen JL, Fisher LJ, Tarnowski JM, Hager MM, Kroc SA, Bertsch NL, Agre KE, Jackson JL, Macklin-Mantia SK, Murphree MI, Rust LM, Summer Bolster JM, Beck SA, Atwal PS, Ellingson MS, Barnett SS, Rasmussen KJ, Lahner CA, Niu Z, Hasadsri L, Ferber MJ, Marcou CA, Clark KJ, Pichurin PN, Deyle DR, Morava-Kozicz E, Gavrilova RH, Dhamija R, Wierenga KJ, Lanpher BC, Babovic-Vuksanovic D, Farrugia G, Schimmenti LA, Stewart AK, Lazaridis KN. Impact of integrated translational research on clinical exome sequencing. Genetics in Medicine. 2021; doi:10.1038/s41436-020-01005-9.
Citations are from PubMed, a service of the U.S. National Library of Medicine. PubMed comprises references and abstracts from MEDLINE, life science journals and online books:
Find more publications authored by Mayo Clinic experts in the area of translational omics.
Featured blog posts
Read about translational omics breakthroughs and meet our program team on the Center for Individualized Medicine blog:
Multimedia
Molecular Genetics and Genomic Medicine: Margot A. Cousin, Ph.D.
Watch Dr. Cousin discuss her Molecular Genetics and Genomic Medicine paper, "Pharmacogenomic findings from clinical whole-exome sequencing of diagnostic odyssey patients."
Genetic Testing's Impact on Patient Care — Paige's Story
Whole-exome sequencing probes into a young patient's bone and joint pain.
Individualized Medicine — Javrie's Story
Obscure symptoms are mapped to a rare pediatric disorder.
A Journey of Hope — Karter's Story
RNA sequencing identifies DNA changes that caused genetic irregularities affecting Karter's growth and development.