New Tools To Visualize Mitochondria
Reconstruction of neural mitochondria
Reconstruction of neural mitochondria
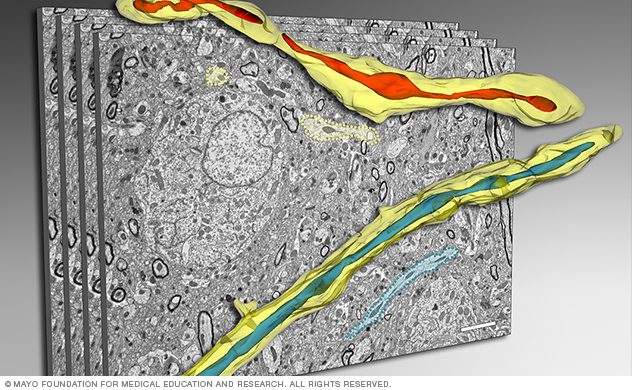
Reconstruction of neural mitochondria
This figure shows a 3D electron microscopy reconstruction of mitochondria in the brain tissue using serial sectioning.
To accurately assess mitochondria morphology in healthy and diseased brain tissue, Dr. Trushina's team collaborated with the Microscopy and Cell Analysis Core to develop a novel 3D electron microscopy (EM) reconstruction technique. This method allows scientists to see 3D details of mitochondrial structure in the context of a 3D human or mouse brain.
Our team also developed conditions that allow tissue preparation and processing free from artifacts often associated with the dynamic nature of mitochondrial morphological changes.
Using these techniques, our lab identified a novel mitochondrial phenotype called mitochondria-on-a-string (MOAS). We also linked MOAS to the fundamental mitochondrial response to stress.
The development of this advanced tool led to multiple collaborations with researchers both at Mayo Clinic and other institutions. Our collaborations focus on the molecular mechanisms of Alzheimer's disease and Parkinson's disease using human and animal model brain tissue.
Detailed visualization of mitochondrial morphology using our 3D electron microscopy reconstruction technique to better understand disease progression could help researchers identify relevant therapeutics.
3-D reconstruction of mitochondrial structure
This video shows an animated 3D reconstruction of mitochondrial structure in the hippocampal tissue of an APP-PS1 mouse at 24 weeks of age.
For 3D reconstruction of the mitochondrial structure, our team began by inverting the grayscale of the individual electron microscopy section images so that the organelles became bright objects. We then sequentially coregistered the inverted images using the Normalized Mutual Information 2D registration program in Analyze, a comprehensive multidimensional medical image processing, visualization and analysis software package developed by the Biomedical Imaging Resource Core. Sequential coregistering is an automated procedure that aligns similar images based on the statistical distribution of paired pixels compared to the distribution in either image alone.
Finally, we rendered the inverted, coregistered stack using maximum intensity projection. Each pixel in the rendered image represents the brightest voxel in a ray from the viewer's eye through the entire stack of sections. This greatly improves the ability to see and understand 3D mitochondrial structure in context.
3-D electron microscopy reconstruction of mitochondria in nontransgenic and APP-PS1 mice
This video shows 3D electron microscopy reconstruction of mitochondria in multiple neuropils in the hippocampi of nontransgenic (left) and APP-PS1 (right) mice.
Mitochondria in Alzheimer's disease mice form MOAS, much longer structures compared to nontransgenic mice. Formation of MOAS could protect mitochondria from degradation, preserving residual functions and promoting cell survival under stress. Two representative sets of standard TEM images of 0.09 μm thickness; from 10 to 40 consecutive serial sections, were taken into reconstruction.