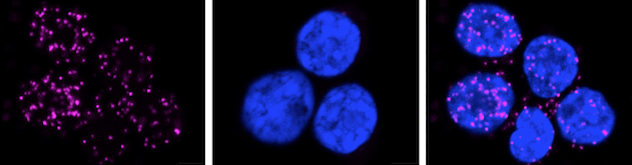
 FOXP3 and EZH2 binding in human Treg cells
FOXP3 and EZH2 binding in human Treg cells
Human T-regulatory (Treg) cells depicting binding of enhancer of zeste homolog 2 (EZH2) and forkhead box protein 3 (FOXP3) via phospholipase A signaling in a proximity ligation assay (PLA) (pink dots). Source: Bamidele AO, et al. Cellular and Molecular Gastroenterology and Hepatology. 2019; doi:10.1016/j.jcmgh.2018.08.009.
Focus Areas
The Immuno-Epigenetics Lab team has several projects studying the pathophysiology of inflammatory bowel disease and how the immune system contributes to intestinal inflammation. These projects aim to characterize the immuno-epigenetics phenotype of inflammatory bowel disease, particularly the effect of environmental pressures on the epigenetics of both epithelial and immune cellular behavior that result in chronic intestinal inflammation.
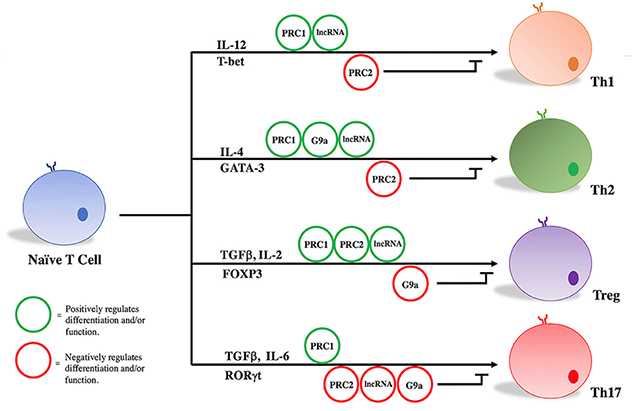 Naive T-cell differentiation
Naive T-cell differentiation
Key transcription factors, epigenetic regulators and cytokines are important components in differentiation and maintenance of T-effector subsets and T-regulatory cell functions. Source: Gaballa JM, et al. Frontiers in Immunology. 2018; doi:10.3389/fimmu.2018.02955.
The team uses human tissue, humanized mouse models and state-of-the-art technologies such as:
- Single-cell RNA sequencing.
- Assay for transposase-accessible chromatin (ATAC).
- Chromatin immunoprecipitation sequencing (ChIP seq).
- 3D chromatin modeling using Hi-ChIP.
Spatial biology technologies validate ex vivo findings in tissue from patients with inflammatory bowel disease.
The lab's environment allows advanced modeling in humanized mouse systems and organoids isolated from the lab's robust inflammatory bowel disease clinical program. Extensive collaboration with Mayo Clinic's Division of Colon and Rectal Surgery, Inflammatory Bowel Disease Clinic and Clinical Trials unit allows facile translation of mechanistic observations to the clinic.
The lab is translating its findings into practice through ex vivo engineering of T cells using epigenetic modifiers and CRISPR-Cas9 molecular engineering.
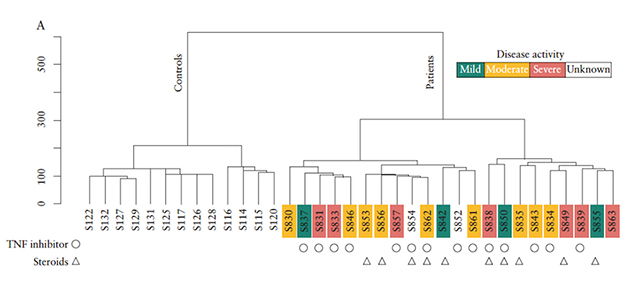 Global lncRNA in CD4+ cells
Global lncRNA in CD4+ cells
Global long noncoding RNAs (lncRNAs) from mucosal CD4+ T cells gene expression separate participants with inflammatory bowel disease from those who are healthy. Unsupervised clustering of lncRNAs differentiates participants with inflammatory bowel disease from healthy controls. Source: Braga-Neto MB, et al. Journal of Crohn's and Colitis. 2020; doi:10.1093/ecco-jcc/jjz109.