The epigenome as a link between the environment and human disease
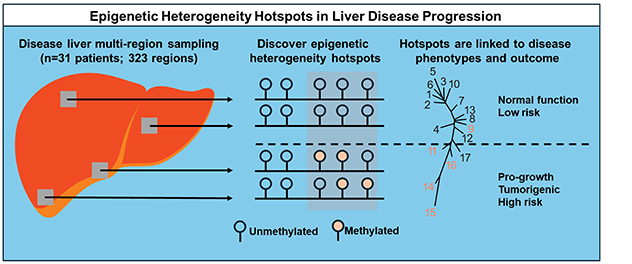 Deciphering epigenetic heterogeneity during liver disease and hepatocellular carcinoma
Deciphering epigenetic heterogeneity during liver disease and hepatocellular carcinoma
Multiple regions of a diseased livers, typically obtained from patients undergoing liver transplants, are sampled for DNA methylation and expression. The lab applies the principles of phylogenetics to discover early epigenetic changes driving liver disease progression and hepatocellular carcinoma.
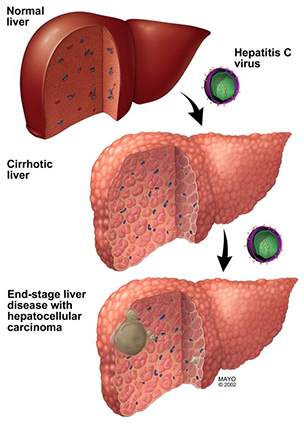 Liver disease progression
Liver disease progression
This diagram elucidates how epigenetic changes due to environmental exposures such as hepatitis C virus infection (shown), or other exposure such as chronic alcohol misuse or metabolic syndrome-associated obesity (not shown), impact the development and progression of liver disease and liver cancer.
Hepatocellular carcinoma is a leading cause of cancer-related death worldwide, and its incidence is increasing in the U.S. Unlike those for many other cancers, the predisposing risk factors for most hepatocellular carcinomas are known. Chronic alcoholism, hepatitis B and C viral infections, and metabolic syndrome driven by obesity account for most of these cancers worldwide.
Hepatocellular carcinoma has a long clinical course and a well-defined precancerous progression. Many of the changes that occur in hepatocytes during the long preneoplastic phase of liver disease, including chronic inflammation, progressing fibrosis and eventually cirrhosis, appear to be epigenetic rather than genetic in nature. This observation likely reflects the known ability of the epigenome to act as an interface with the environment.
External insults such as viral infection first alter the epigenome, leading to maladaptive changes in gene expression that promote further epigenetic, and eventually genetic, damage. Alterations in immune cell activation and the extracellular milieu in the liver also likely contribute to this damage.
Studies in the Epigenetic Etiology of Human Disease Laboratory focus on roles for the DNA epigenetic marks in hepatocellular carcinoma initiation and progression and how these agents deregulate enhancer function and create aberrant epigenetic states consistent with an elevated risk of cancer formation. Epigenetic marks and expression patterns are interrogated across the entire genome of primary human liver disease samples. These data are related to available patient clinical-pathological data and cell line and organoid model systems.
To further understand how epigenetic changes drive liver disease and hepatocellular carcinoma, the laboratory is investigating spatial epigenetic heterogeneity within diseased and neoplastic regions of the liver. The lab also is working to distinguish between epigenetic changes that are drivers and those that are passengers, also called late-stage events.
To accomplish this, we use whole livers obtained through liver transplant for end-stage liver disease or early hepatocellular carcinoma, sample throughout the 3D structure of the tissue, and examine DNA methylation and transcriptional heterogeneity. Findings are correlated with clinical data, gene expression and immunostaining and used in phyloepigenetic analyses to define epigenetic changes that act as early driving events. Such analyses are expected to yield novel driver events for hepatocellular carcinoma, new therapeutic targets and, potentially, early-disease biomarkers.
The laboratory also is working to understand the long-term ramifications of chronic hepatitis C virus infection of the liver followed by sustained virologic remission or cure with the direct-acting antiviral drugs that became available in 2011. Viral cure is now highly efficient and although long-term risk of hepatocellular carcinoma driven by hepatitis C virus infection is reduced, it is not eliminated. This necessitates long-term screening of patients.
The Epigenetic Etiology of Human Disease Laboratory is testing whether chronic hepatitis C virus infection leaves an epigenetic memory, also called a scar, after viral cure. Such a scar may elevate the risk of liver disease progression.
Epigenetic scars might manifest in the form of DNA damage, a proinflammatory phenotype or altered cellular responses to stress, or aberrant responses to pro-cell death signals. Using models of hepatitis C virus infection and viral clearance in cell culture supported by analysis of patient liver tissue, we aim to understand:
- How the virus alters the epigenome.
- The role of innate immune pathways.
- How the epigenome and cellular growth phenotypes respond to viral clearance.
Interestingly, hepatitis C virus infection — which itself induces a cellular interferon response in an often-futile attempt to clear the virus — drives many epigenetic and transcriptional changes that mirror those of interferon-alpha treatment. Therefore, we are testing whether chronic interferon-alpha exposure is a key player underlying the cellular responses to hepatitis C virus infection. Our longer term goal is to identify ways to further reduce cancer risk in people cured of chronic hepatitis C virus infection. This may be feasible given the reversible nature of epigenetic marks.