 Understanding and targeting epigenetic changes driving human disease and cancer
Understanding and targeting epigenetic changes driving human disease and cancer
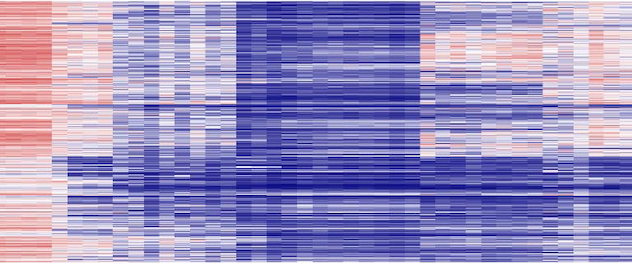
The Epigenetic Etiology of Human Disease Laboratory led by Keith D. Robertson, Ph.D., at Mayo Clinic is working to understand how epigenetic modifications are disrupted during cancer initiation and progression, how detrimental environmental exposures work through the epigenome to promote disease, and how germline mutations in epigenetic regulators lead to neurodevelopmental conditions. This heat map (visualization tool) illustrates changes in epigenetic marks across the genome from a panel of primary renal cell cancers.
Overview
 Epigenetic marks and their effects on transcription and genome packaging
Epigenetic marks and their effects on transcription and genome packaging
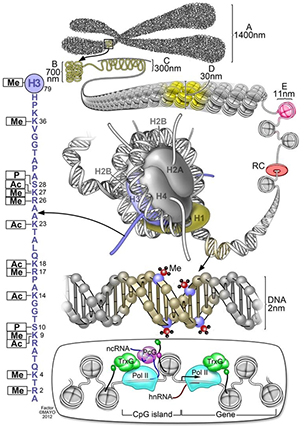
This schematic summarizes different classes of epigenetic marks, including methylation on DNA, the wide array of histone post-translational modifications on the histone octamer (with a blowup of modifications to the histone H3 N-terminal tail shown on the left) and the binding of various epigenetic regulatory proteins (bottom). Me stands for methylation. P stands for phosphorylation. And Ac stands for acetylation.
The primary focus of the Epigenetic Etiology of Human Disease Laboratory led by Keith D. Robertson, Ph.D., is to understand the role of epigenetics, particularly DNA and histone methylation, in human health and disease.
Epigenetic modifications regulate the accessibility and stability of DNA and ultimately determine how, when and where genetic information is used to specify cellular identity without altering its primary sequence. Epigenetic marks within the human genome include those that target the DNA, such as methylation and hydroxymethylation. Epigenetic marks also include those that target the histone proteins that package the DNA, such as lysine methylation and acetylation. Methylation also modifies RNA at several bases.
A common theme of epigenetic modifications is their regulation and reversibility by a three-component system:
- Writers. These enzymes put on the mark.
- Readers. These proteins bind the mark and help carry out its functions.
- Erasers. This group of proteins removes the mark.
Regulated changes in epigenetic marks are essential for normal human development and maintenance of cellular identity. Unique epigenetic profiles enable stem cells to differentiate into any cell type despite their DNA content being identical to specialized cell types throughout the body. Similarly, the unique placement of epigenetic marks that participate in the regulation of cell type-specific patterns of gene expression define most differentiated cell types within the human body.
In human diseases such as cancer, diabetes, cardiovascular disease and neurological conditions, epigenetic marks are subtly to profoundly altered, typically in disease- and stage-specific manners. Sometimes genes that regulate the epigenome become disrupted early during development. While these changes may not dramatically alter cancer predisposition, they contribute to several rare neurodevelopmental and overgrowth and undergrowth syndromes.
 DNA cytosine modification cycle in mammalian cells
DNA cytosine modification cycle in mammalian cells
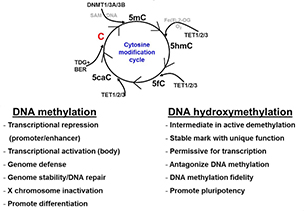
This diagram shows the relationships between the four DNA epigenetic modifications and their regulators. DNMT stands for DNA methyltransferase. TET stands for ten-eleven translocation protein.
For example, disrupted DNA methylation patterns are a universal feature of cancer cells. They occur early during tumor development, and they actively contribute to cancer initiation and progression. The epigenome is thought to act as an interface between factors in the highly variable environment and the static genome. Nutrition, viral infection, stress and inflammation are just a few of the factors that can alter the epigenome.
In this capacity, the epigenome takes inputs from environmental cues and modulates use, also called expression, of the genome. This modulation could be a positive adaptive response that promotes cellular fitness or it could be a change that results in pathological gene expression that predisposes people to elevated disease risk.
A growing number of drugs target aberrant DNA and histone epigenetic changes. These drugs are becoming available for patient treatment, clinical trials and laboratory studies. They provide novel ways to target tumor cells by restoring normal epigenetic states. That's because epigenetic modifications, unlike genetic mutations, are reversible as part of their normal biology. The use of such drugs can be tailored to a person's individual epigenetic and genetic landscape, delivering personalized medicine.
In addition, detecting aberrant DNA methylation in DNA derived from bodily fluids such as blood plasma, urine or stool holds great promise as a source of noninvasive early detection biomarkers. For example, disease-specific DNA methylation signatures within cell-free DNA circulating in plasma and released from diseased tissue have emerged as a powerful source of early detection and prognostic biomarkers for cancer and diseases not related to cancer.
Since this DNA retains epigenetic marks characteristic of its cell of origin, epigenetic biomarkers can identify not only the presence of disease but also the affected tissue or cell type. This permits healthcare professionals to identify people most at risk and begin treating them while the disease is in its earliest and most treatable stage.
Projects
The lab's projects focus on studying epigenetic marks in normal cells, understanding how these processes become disrupted during cellular transformation or development, and determining how we can take advantage of disease-specific epigenetic changes for novel biomarkers and therapeutic targets.
Affiliations
Dr. Robertson's Epigenetic Etiology of Human Disease Laboratory works closely with these Mayo Clinic research groups and organizations.