 Genetic screening platform
Genetic screening platform
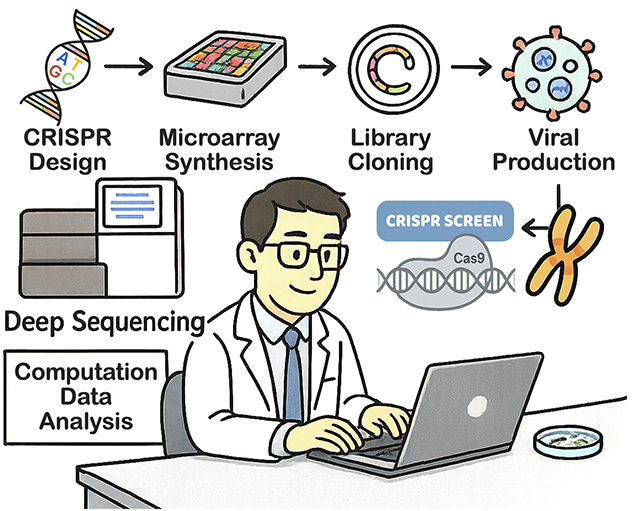
Dr. Chen's lab integrates CRISPR library design, high-throughput screening and computational analysis to uncover genetic mechanisms driving cellular function and disease.
Research
CRISPR gene tiling technology for de novo functional element discovery
We have developed a high-resolution genetic screening approach called single-cell CRISPR gene tiling, also known as sc-tiling. This method enables the de novo identification of functional elements within proteins by performing saturation mutagenesis through CRISPR-mediated genome editing.
Our custom in-house screening pipeline integrates:
- Microarray-based oligo synthesis.
- Single-guide RNA (sgRNA) library cloning.
- Lentiviral production and transduction.
- Drug sensitivity and resistance screening.
- 10x Genomics CRISPR capture and single-cell RNA sequencing.
- Comprehensive data analysis.
We envision that this technology will establish a new research frontier at the intersection of functional genomics and protein domain analysis, ultimately facilitating the discovery of novel therapeutic pockets.
Publication
Yang L, Chan AKN, Miyashita K, Delaney CD, Wang X, Li H, Pokharel SP, Li S, Li M, Xu X, Lu W, Liu Q, Mattson N, Chen KY, Wang J, Yuan YC, Horne D, Rosen ST, Soto-Feliciano Y, Feng Z, Hoshii T, Xiao G, Muschen M, Chen J, Armstrong SA, Chen CW. High-resolution characterization of gene function using single-cell CRISPR tiling screen. Nature Communications. 2021; doi:10.1038/s41467-021-24324-0.
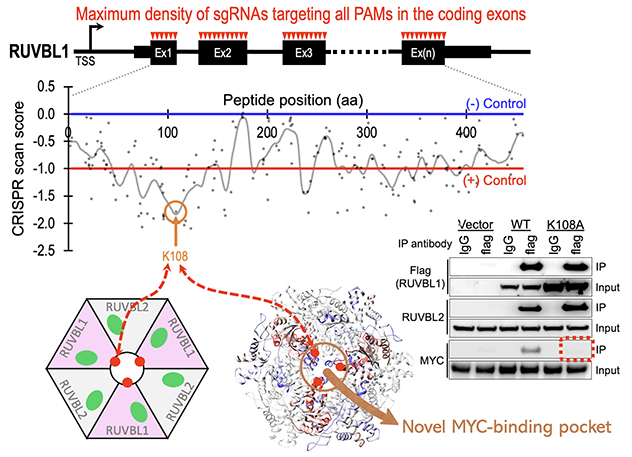 CRISPR tiling reveals a novel MYC-binding pocket in RUVBL1
CRISPR tiling reveals a novel MYC-binding pocket in RUVBL1
A high-resolution CRISPR tiling screen of RUVBL1 identified lysine 108 (K108) as a critical residue for binding the oncogenic MYC protein and promoting Ewing sarcoma pathogenesis.
Novel mechanism of MYC chromatin targeting
Ewing sarcoma is a pediatric cancer characterized by aggressive tumor growth. Through an epigenetics-focused CRISPR screen with about 3,600 sgRNAs targeting approximately 720 epigenetic regulators, we identified the NuA4 histone acetyltransferase complex as essential for Ewing sarcoma maintenance.
Our data highlight RUVBL1, a key component of the NuA4 complex, as a master regulator of MYC chromatin targeting and transcriptional activation. Using our CRISPR tiling pipeline, we further dissect the critical position of RUVBL1, mediating its interaction with MYC. We anticipate that these findings will guide the development of novel therapeutics to enhance MYC-targeted treatments across multiple cancer types.
Publication
Li M, Yang L, Chan AKN, Pokharel SP, Liu Q, Mattson N, Xu X, Chang WH, Miyashita K, Singh P, Zhang L, Li M, Wu J, Wang J, Chen B, Chan LN, Lee J, Zhang XH, Rosen ST, Muschen M, Qi J, Chen J, Hiom K, Bishop AJR, Chen CW. Epigenetic control of translation checkpoint and tumor progression via RUVBL1-EEF1A1 axis. Advanced Science. 2023; doi:10.1002/advs.202206584.
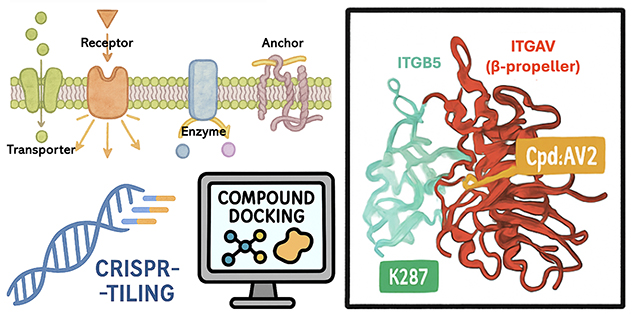 CRISPR-TICA: A CRISPR-tiling-instructed computer-aided pipeline for drug discovery
CRISPR-TICA: A CRISPR-tiling-instructed computer-aided pipeline for drug discovery
By integrating cell surface proteome screening, CRISPR tiling and in silico compound docking, we developed a novel pipeline to identify druggable surface pockets on ITGAV, advancing next-generation cancer therapeutics.
Novel drug discovery pipeline informed by CRISPR gene tiling
Using a CRISPR library targeting the cell surface proteome of about 3,000 sgRNAs across approximately 580 proteins, we identified integrin alpha-V (ITGAV), an extracellular matrix receptor, as a top essential cell surface protein in multiple cancer types. We mapped a functionally critical surface pocket on ITGAV suitable for therapeutic targeting by applying CRISPR gene tiling.
By integrating this approach with compound docking simulations, we developed a novel drug discovery workflow, CRISPR-tiling instructed computer-aided (CRISPR-TICA). This study marks a significant step forward in next-generation therapeutic development and lays the foundation for innovative drug discovery strategies.
Publication
Mattson NM, Chan AKN, Miyashita K, Mukhaleva E, Chang WH, Yang L, Ma N, Wang Y, Pokharel SP, Li M, Liu Q, Xu X, Chen R, Singh P, Zhang L, Elsayed Z, Chen B, Keen D, Pirrotte P, Rosen ST, Chen J, LaBarge MA, Shively JE, Vaidehi N, Rockne RC, Feng M, Chen CW. A novel class of inhibitors that disrupts the stability of integrin heterodimers identified by CRISPR-tiling-instructed genetic screens. Nature Structural & Molecular Biology. 2024; doi:10.1038/s41594-024-01211-y.
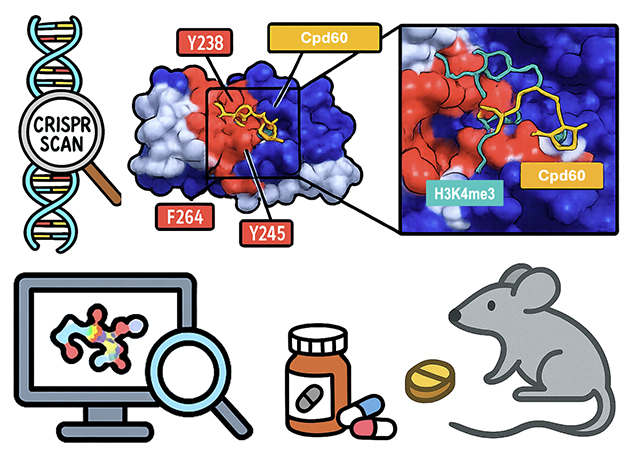 Uncovering a druggable Tudor domain in leukemia.
Uncovering a druggable Tudor domain in leukemia.
A CRISPR-based scan of SGF29 led to the discovery of a Tudor domain small-molecule inhibitor Cpd60 through integrated genetic, structural and computational approaches.
Novel epigenetic targeting in leukemia
Epigenetic dysregulation frequently drives MLL-rearranged (MLL-r) leukemias, making the epigenetic machinery a promising therapeutic target. To explore this, we conducted a CRISPR screen using 1,000 sgRNAs targeting the exon regions of all 59 known Tudor domains — chromatin reader modules that recognize methylated lysine and arginine residues on histones. This screen identified SGF29 as a novel essential gene in MLL-r leukemia.
By integrating CRISPR-tiling, 3D structural modeling and in silico compound docking, we identified the first small-molecule inhibitor of SGF29 capable of suppressing MLL-r leukemia cells. We anticipate that this multidisciplinary strategy will provide a streamlined framework for therapeutic discovery, integrating genetic, functional, structural and chemical insights.
Publication
Chan AKN, Han L, Delaney CD, Wang X, Mukhaleva E, Li M, Yang L, Pokharel SP, Mattson N, Garcia M, Wang B, Xu X, Zhang L, Singh P, Elsayed Z, Chen R, Kuang B, Wang J, Yuan YC, Chen B, Chan LN, Rosen ST, Horne D, Muschen M, Chen J, Vaidehi N, Armstrong SA, Su R, Chen CW. Therapeutic targeting Tudor domains in leukemia via CRISPR-scan assisted drug discovery. Science Advances. 2024; doi:10.1126/sciadv.adk3127.
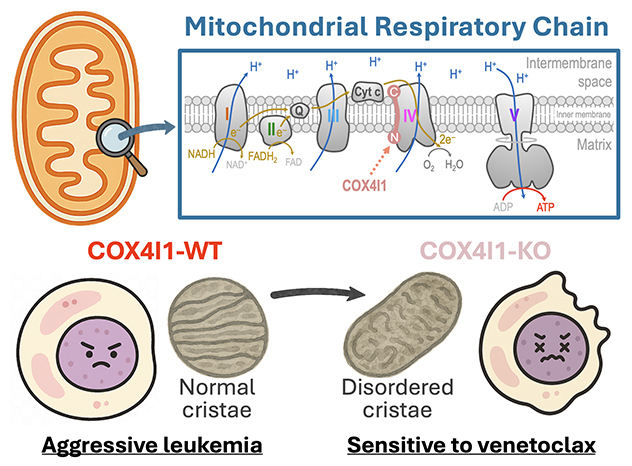 Targeting mitochondrial homeostasis in leukemia
Targeting mitochondrial homeostasis in leukemia
COX4I1 regulates the assembly of complex IV (cytochrome c oxidase) and preserves normal mitochondrial cristae ultrastructure. Inhibiting COX4I1 enhances the sensitivity of leukemia cells to venetoclax treatment.
Mitochondrial pathway regulating therapeutic sensitivity in leukemia
Using a cell signaling-focused CRISPR library of about 3,500 sgRNAs targeting approximately 420 signaling mediators, we identified COX4I1 — a nuclear-encoded subunit of cytochrome c oxidase — as an essential factor in leukemia. To elucidate its role, we employed a multi-omics approach combining transcriptomics, metabolomics, proteomics, CRISPR tiling-based structural genetics and in vivo mouse modeling.
Our findings reveal that COX4I1 regulates mitochondrial homeostasis, cristae architecture, oxidative phosphorylation and sensitivity to venetoclax. This study uncovers a novel mitochondrial mechanism underlying therapeutic response and proposes a new strategy for combination therapy in leukemia treatment.
Publication
Zhang L, Zhang H, Wang TY, Li M, Chan AKN, Kang H, Foong LC, Liu Q, Pokharel SP, Mattson NM, Singh P, Elsayed Z, Kuang B, Wang X, Rosen ST, Chen J, Yang L, Chou TF, Su R, Chen CD. Nuclear control of mitochondrial homeostasis and venetoclax efficacy in AML via COX4I1. Advanced Science. 2025; doi:10.1002/advs.202404620.
All publications
View all publications for David (Chun-Wei) D. Chen, Ph.D.